COVID-19 GPH: tracking the contribution of genomics and precision health to the COVID-19 pandemic response
The COVID-19 pandemic, caused by SARS-CoV-2, broke out at the start of 2020 [1]. The global scientific community responded with extraordinary effort, sharing information in online databases, preprints, and scientific publications. Based on PubMed and preprint server searches, more than 200,000 scientific articles and preprints were published during two years of the pandemic. They report the results of basic, clinical, and population-based investigations, ranging from studies of the virus itself to the global impact of the pandemic on health, economics, and daily life. Rapid growth of the scientific literature on COVID-19 makes it difficult for scientists, clinical and public health professionals, and the community in general to keep up databases such as LitCovid [2] and the World Health Organization COVID-19 database [3], are a key resource for researchers, policy-makers, and the public.
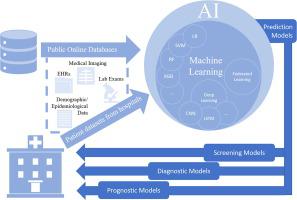
Genomics and data science—including computational methods often referred to as machine learning or artificial intelligence—have been instrumental in many aspects of research on COVID-19. These methods have provided insights into SARS-CoV-2 and how it evolves and spreads in populations, as well as susceptibility to COVID-19 infection, risk of severe outcomes, and role of COVID-19 treatments. Genomic surveillance has documented the emergence and spread of the omicron and delta SARS-CoV-2 variants in Denmark [4] as well as the unique mutations that differ between these two variants [5]. Other studies have described the role of human genetic polymorphisms in COVID-19 susceptibility [6], the upregulation of proinflammatory cytokine genes in severe COVID-19 patients [7], and the toxicity profiles of 90 possible COVID-19 treatments using machine learning [8].
To track and provide easier access to the application of genomics and precision health in the COVID-19 response, the CDC Office of Genomics and Precision Public Health launched the COVID-19 Genomics and Precision Health knowledge management system and database (COVID-19 GPH) on April 1, 2020. COVID-19 GPH is a component of the Public Health Genomics and Precision Health Knowledge Base (PHGKB). PHGKB features a suite of curated and continuously updated, searchable databases of published scientific literature, CDC resources, and other materials that address the translation of genomics and precision health discoveries into improved health care and disease prevention [9]. Two databases that capture the broad spectrum of biomedical research on COVID-19 have been established by separate groups at the US National Institutes of Health (NIH): LitCOVID at the National Library of Medicine [2], and the iSearch COVID-19 Portfolio at the Office of Portfolio Analysis [10]. In contrast to these databases, COVID-19 GPH was developed to select a subset of the technology-intense scientific literature on COVID-19 that is most relevant to public health and population medicine. Because COVID-19 GPH is curated, users can quickly identify information related to genomics and precision public health without having to compose a complex search query. COVID-19 GPH also links to news, reports, and other relevant information from CDC, NIH and other public health organizations, all updated daily. Thus, in addition to a searchable archive of scientific literature, COVID-19 GPH offers an easily accessible, online update that helps users keep abreast of the latest developments. Here we describe this unique database and its contribution to organizing the rapidly expanding knowledge base on COVID-19.
Construction and content
Implementation
COVID-19 GPH is a web-based application based on J2EE technology [11] with Java open-source frameworks including Hibernate [12] and Strut [13]. As a component of the PHGKB system, COVID-19 GPH has been built on and integrated into the overall architecture of PHGKB described previously [14, 15].
Data retrieval
Data are collected mainly from PubMed, the NIH iSearch COVID-19 Portfolio [10], LitCovid [2], and common media sources by an automatic retrieval and text mining strategy [15], combined with manual curation by domain experts at the Centers for Disease Control and Prevention (CDC) (Fig. 1). Data are retrieved by four main approaches. First, the scientific publications are retrieved from PubMed daily by an automated script using NCBI Eutils [16] using two specifically designed queries (Additional file 1: Appendix I). Second, we use the same queries to search the NIH iSearch COVID-19 Portfolio website and download records retrieved in spreadsheet format which are subsequently uploaded to the database using an automatic script. Third, we automatically retrieve records classified to the epidemic forecasting category in the LitCovid database using the LitCovid RSS feed. Finally, CDC staff selects online news and other reports from our weekly horizon scan for the Genomics Health Impact Update [17] and Advanced Molecular Detection Clips [18] and other sources. The inclusion and exclusion criteria for these weekly scans are described in detail in the Additional file 1: Appendix II. The curation pipelines include a series of computer scripts for scheduled automatic data retrieval and uploading, along with a web-based curation interface that CDC domain experts use to select and curate important news, reports, and articles. The PubTator web service is used to annotate gene information in PubMed records. A text mining technique [14] is used to identify and standardize the country information associated with the authors in PubMed records. All data selection processes are performed daily. To prevent potential record duplication through multiple retrieval processes, we use a de-duplication mechanism based on unique PubMed IDs or publication titles.
Data classification
Data are classified into two main groups: Genomics Precision Health and Non-Genomics Precision Health (Additional file 1: Appendix II). They are then further classified automatically into 10 different categories: eight based on the PubTator [19] classifier in the LitCovid database [2] (mechanism, treatment, prevention, diagnosis, forecasting, surveillance, transmission) by querying and parsing LitCovid RSS feeds, and three created using text mining scripts (vaccine, variant, health equity) using keyword searching (keywords in Additional file 1: Appendix III). Data are also classified to 12 topics with their own sub-databases in PHGKB (Cancer; Diabetes; Heart, Lung, Blood and Sleep Diseases; Rare Diseases; Health Equity; Family Health History; Reproductive and Child Health; Pharmacogenomics; Neurological Disorders; Primary Immune Deficiency; Environmental Health).
Evaluation of data retrieval performance
To validate our automated data retrieval process, we generated a 499-item random sample from the LitCovid database on April 23, 2021. These records were screened automatically as shown in Fig. 1 and classified as positive (included in the database) or negative (excluded from the database). The automatic query included 55 articles and excluded 444 articles. At the same time, two domain experts independently reviewed the same 499 records manually and classified them according to the database inclusion and exclusion criteria. They discussed all 23 instances of disagreement and arrived at a final classification by consensus. The experts included 50 articles and excluded 449 articles. The performance of the automated retrieval process was evaluated by calculating its specificity and sensitivity, using expert classification as the gold standard. The automatic curation process has an estimated sensitivity of 0.82 and specificity of 0.97 for PubMed articles (Table 1).
User interface and features
The COVID-19 GPH web-based user interface is shown in Fig. 2. The landing page of the site provides two main sections that list important publications picked by a CDC domain expert (Spotlight) and the most recent records added to the database (Latest News and Publications). Summary statistics are on the left side of the page. The user interface allows users to perform a free text search on any topic. The search results can be further stratified by five filters (Country, Journal, Gene, Publication Type and Publication Category). The filtering process can be repeated until a desired search result is achieved. Users can also perform a search on sub-datasets for 10 special topics in PHGKB. Two graphs can be drawn dynamically to summarize the search results: (1) Distribution of Publications by Month and (2) Distribution of Publication by Category. Users also can sign up for a COVID-19 GPH Weekly Update email newsletter that includes COVID-19 related items selected by CDC staff in these categories: Pathogen and Human Genomics Studies, Non-Genomics Precision Health Studies and News/Reviews/Commentaries.
Utility and discussion
Description
COVID-19 GPH is an open access, online database containing links to original studies, reviews, commentaries, and news relevant to genomics, machine learning, or the use of big data in COVID-19 research. Although most records are extracted from PubMed, the database also contains preprints as well as selected online news, reports, and publications (Table 2). Included articles reference 845 human genes, with ACE2 being the most common.
The database contains information on the surveillance, investigation, diagnosis, treatment, prevention, and control of COVID-19. The contents are divided into two main sections, Genomics Precision Health (GPH) and Non-Genomics Precision Health (Non-GPH). GPH contains literature focused on applications of pathogen and human genomics. The literature in Non-GPH relates to the use of big data, data science, digital health, machine learning, predictive analytics and forecasting methods. As of February 11, 2022, the database contains 31,597 articles (22,597 GPH, 9,000 Non-GPH). Articles in both categories may be classified into one or more of 11 publication categories (Table 3). These categories are not mutually exclusive, and an article may be assigned to more than one. In the entire database, the largest category is “Variants” (n = 6735) and the smallest is “Health Equity” (n = 804); however, the relative sizes of these categories differ between the GPH and non-GPH groups (Fig. 3). Some common topics among articles included in the database are listed in Table 4, along with examples [16,17,18,19,20,21,22,23,24,25,26,27,28,29]. We estimated the fraction of scientific literature on selected for COVID-19 GPH by dividing the number of PubMed records in COVID-19 GPH by the number of PubMed records in LitCovid: 22983/221241 (10%) based on the data retrieved on February 11, 2022.
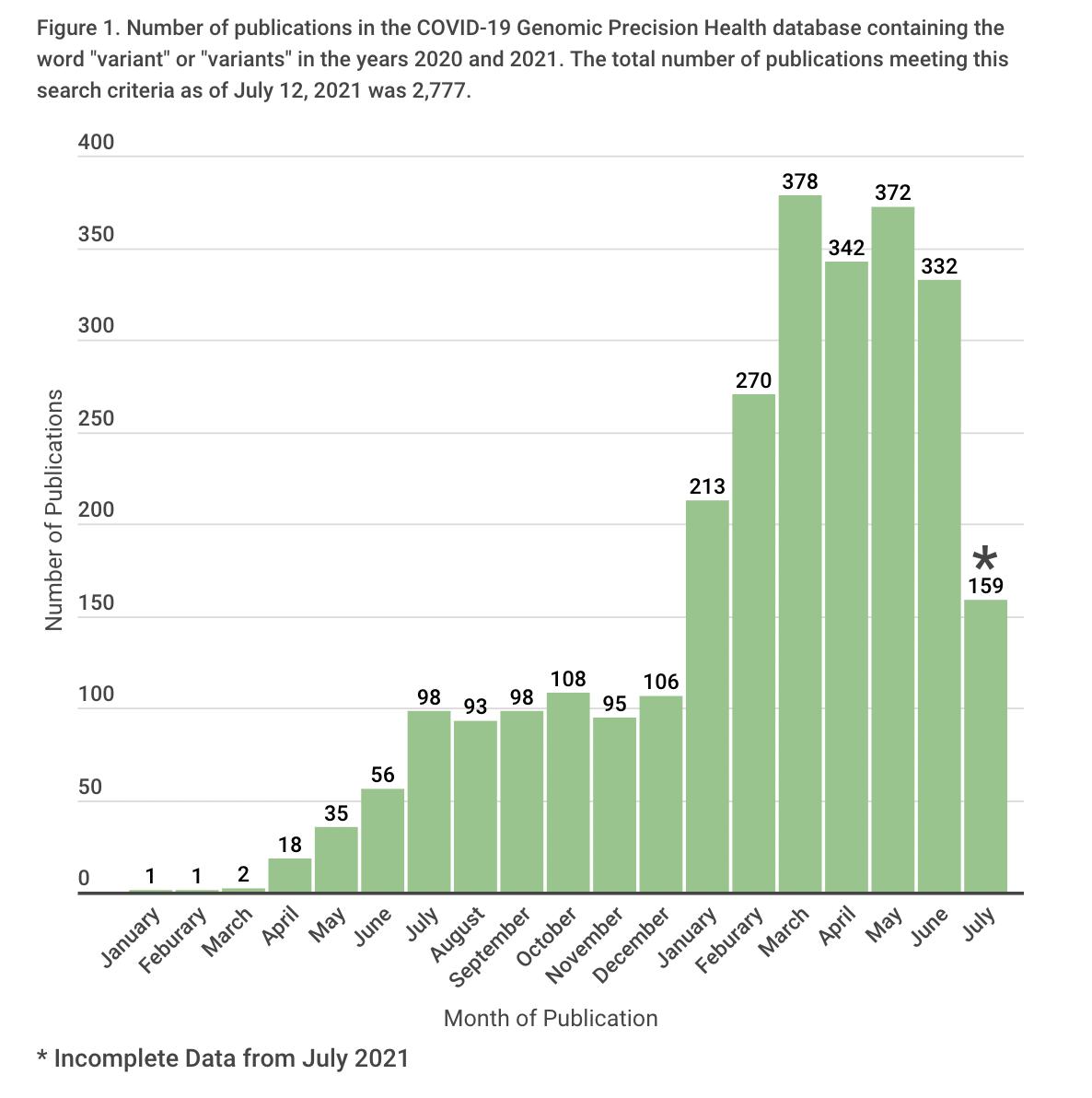
The database can be used to analyze publication trends by month (Fig. 4). After increasing rapidly in early 2020, the number of articles published per month has generally remained between 1000 and 1700. (Note that because of processing time at PubMed, the number for January 2022 may be incomplete). Trends by category tend to be consistent overall, except for prevention and forecasting which peaked 2020 (Fig. 5). Articles in several other categories (variants, vaccine, mechanism, and diagnosis) generally increased in 2021 (Fig. 5).
For each PubMed publication, the database also captures the Altmetric score, a numerical value indicating the amount of attention an article has received [30]. Of the articles with the top 100 Altemetric scores, the vaccine category accounted for the largest share (26%) and the variant category was second (12%) (Fig. 6).
The database simplifies the search for COVID-19 and certain rare diseases, including articles related to 471 of the approximately 7,000 rare diseases on the NIH Genetic and Rare Diseases Information Center website [31]. Users can also search for articles common to COVID-19 GPH and other specialized PHGKB databases. Of the specialized databases, rare disease has the most overlap, 6811 articles, with COVID-19 GPH while Family Health History shares the least number of articles, 10 (Table 5).